From Manual Analysis to Automated Insight: Building a Healthcare AI Taxonomy Pipeline
Introduction
The integration of artificial intelligence into healthcare represents one of the most transformative technological shifts in modern medicine. As AI applications proliferate across medical specialities, healthcare practitioners face an increasingly complex landscape of tools, technologies, and approaches. Understanding this landscape—what applications exist, how they work, and their potential impact on clinical practice—has become crucial for medical professionals seeking to leverage AI effectively.
Traditional approaches to mapping this terrain often rely on manual literature reviews, expert opinion, and theoretical frameworks. While valuable, these methods struggle to keep pace with the rapid evolution of AI in healthcare. They are time-intensive, difficult to update, and often reflect the biases and limitations of their creators.
This article presents an alternative approach: a systematic methodology for developing and validating a practitioner-focused taxonomy of AI applications in healthcare. We begin by presenting a comprehensive taxonomy organised by medical specialities, developed through AI’s deep research of the medical literature. We then outline a vision for automating this taxonomy development process, creating a pipeline that can systematically generate, validate, and visualise taxonomies not only for healthcare AI but potentially for any domain of interest.
Using our healthcare AI taxonomy as a case study, we demonstrate how this automated approach could transform knowledge organisation, making it more systematic, evidence-based, and adaptable to rapid technological change. By bridging the gap between manual analysis and automated insight, we aim to provide a framework that helps practitioners navigate complex technological landscapes more effectively.
The Healthcare AI Taxonomy: A Case Study
A Practitioner-Focused Framework
Understanding the landscape of AI in healthcare presents a significant challenge for practitioners. The rapid proliferation of AI applications across medical domains creates a complex environment that’s difficult to navigate without a structured framework. To address this need, we developed a comprehensive taxonomy that organises AI applications by medical specialty—aligning with how healthcare professionals typically identify and seek information.
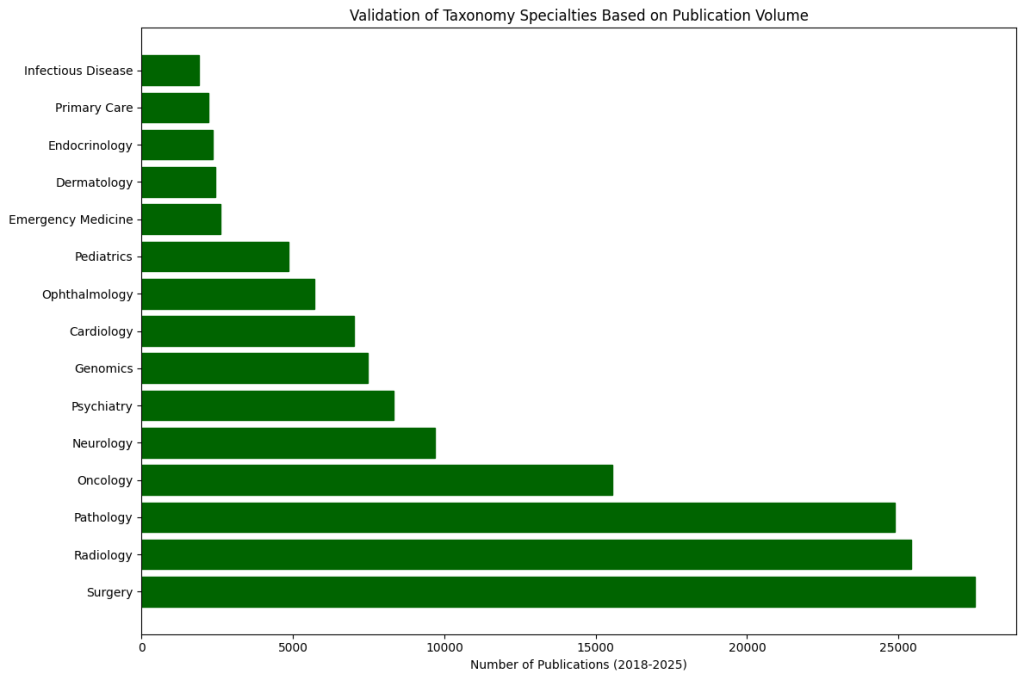
This taxonomy comprises 16 medical specialities with significant AI integration, including Cardiology, Radiology, Ophthalmology, Pathology, Surgery, Genomics, Dermatology, and others. Within each speciality, we identified specific clinical applications where AI is making a meaningful impact, along with the technologies employed and representative research examples.

Taxonomy Structure and Insights
The primary organisation by medical speciality creates an intuitive entry point for practitioners seeking to understand AI applications relevant to their field. For example, a cardiologist can quickly locate applications such as arrhythmia detection, cardiac imaging analysis, cardiovascular risk prediction, ECG interpretation, and periprocedural planning.
Our research revealed several key insights about the current state of AI in healthcare:
- Varied adoption across specialities: Fields rich in imaging data (radiology, pathology, ophthalmology) show more advanced AI integration, while other areas are in earlier adoption stages.
- Common cross-cutting applications: Several application types appear across multiple specialities, including image analysis, risk prediction, workflow optimisation, diagnostic assistance, and personalised treatment planning.
- Evolution of AI roles: Applications are evolving from purely assistive tools to more sophisticated systems that augment clinical decision-making and, in some cases, automate specific tasks.
- Increasing clinical implementation: There is a gradual shift from theoretical research to real-world clinical applications, with growing regulatory approval and integration into clinical workflows.
Development Methodology
This taxonomy was created through a systematic, multi-step process:
- Deep research: Analysing the medical literature to identify AI applications across specialities using Deep Research feature
- Initial categorisation: Organising applications into a two-level hierarchy by speciality
Prompt:
"Search PubMed and create a comprehensive two-level taxonomy of AI applications in healthcare.
The first level should be organized by medical specialties (e.g., Cardiology, Radiology, etc.).
The second level should identify specific AI applications within each specialty.
For each second-level category, provide:
1. The type of AI technology typically used
2. A brief description of how it's applied in clinical practice
3. One example of a representative research paper
Focus on applications that would be most relevant to practicing clinicians."
- Validation: Confirming applications through publication metrics and clinical implementation evidence
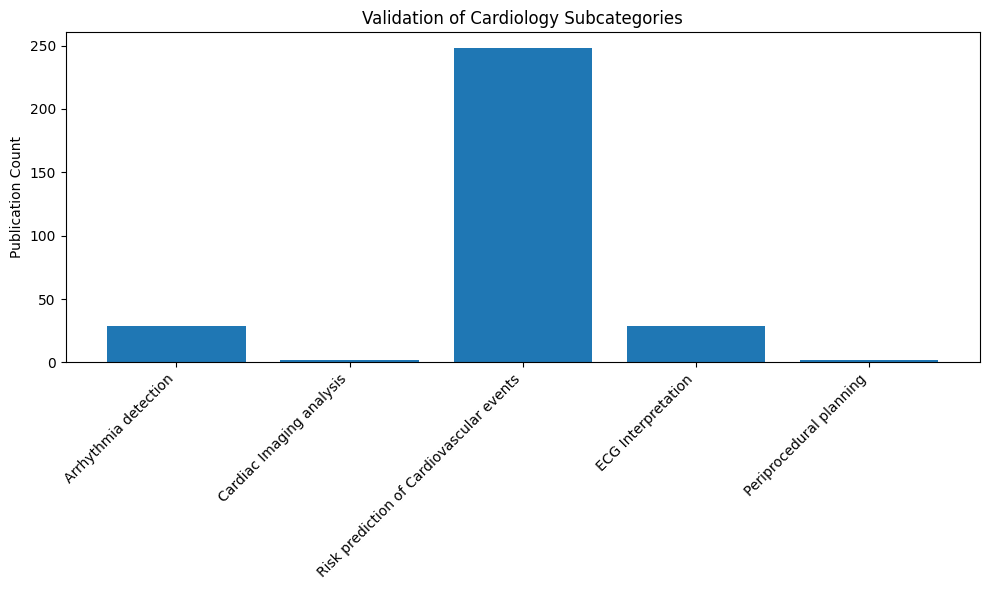
- Refinement: Iteratively improving the taxonomy structure and content
- Visualisationcategorisation: Creating visual representations(infographics) of the taxonomy
While comprehensive and evidence-based, this manual approach required significant time and expertise. It also faces challenges in keeping pace with the rapidly evolving field of AI in healthcare. This limitation prompted us to explore how the taxonomy development process itself might be automated.
Limitations of Manual Taxonomy Development
The manual process of creating our healthcare AI taxonomy, while thorough, revealed several inherent limitations:
- Updating challenges: As new AI applications emerge daily, keeping the taxonomy current would require constant manual monitoring and updates.
- Scalability constraints: Expanding to additional domains or creating similar taxonomies for other fields would multiply the required effort linearly.
- Consistency issues: Manual categorisation decisions can vary based on the researcher’s judgment, creating potential inconsistencies.
Recognising these challenges, we envisioned an automated pipeline that could transform the taxonomy development process. This approach would leverage the strengths of AI-powered research tools and data analytics to create a more efficient, consistent, and updatable taxonomy generation system.
The Opportunity for Automation: A Six-Step Process
To translate our vision into a practical implementation, we designed a six-step pipeline that systematically transforms unstructured knowledge into organised taxonomies. This pipeline combines AI-powered research, data-driven validation, and automated visualisation to create a comprehensive taxonomy development system.
Each step in the pipeline addresses specific aspects of the taxonomy creation process:
.png?raw=true)
Step 1: Initial Query and Data Collection
- Input: Domain or field of interest (e.g., “AI in Healthcare”)
- Process: Generate structured prompts for AI systems to collect relevant information
- Output: Initial search results and topic clustering
Step 2: Taxonomy Structure Generation
- Input: Results from the deep research
- Process: AI analysis to identify hierarchical relationships
- Output: JSON schema with proposed taxonomy structure
Step 3: Literature Validation
- Input: Taxonomy structure JSON
- Process: Automated searches of publication databases (e.g., PubMed) for each category
- Output: Validation data including publication counts and growth trends
Step 4: Taxonomy Refinement
- Input: Initial taxonomy and validation data
- Process: AI analysis to identify gaps, overlaps, and refine categories
- Output: Refined taxonomy JSON structure
Step 5: Visualisation Generation
- Input: Refined taxonomy JSON
- Process: Automated generation of visual representations
- Output: Visual formats such as mindmaps, infographics, and interactive diagrams
Step 6: Documentation Production
- Input: Taxonomy JSON and visualisation files
- Process: AI-generated summary reports
- Output: Comprehensive documentation of the taxonomy
Technical Implementation
We have begun implementation of this pipeline, focusing initially on Steps 1-3 to demonstrate the core concept. Our current approach uses a combination of AI language models and publication database APIs.
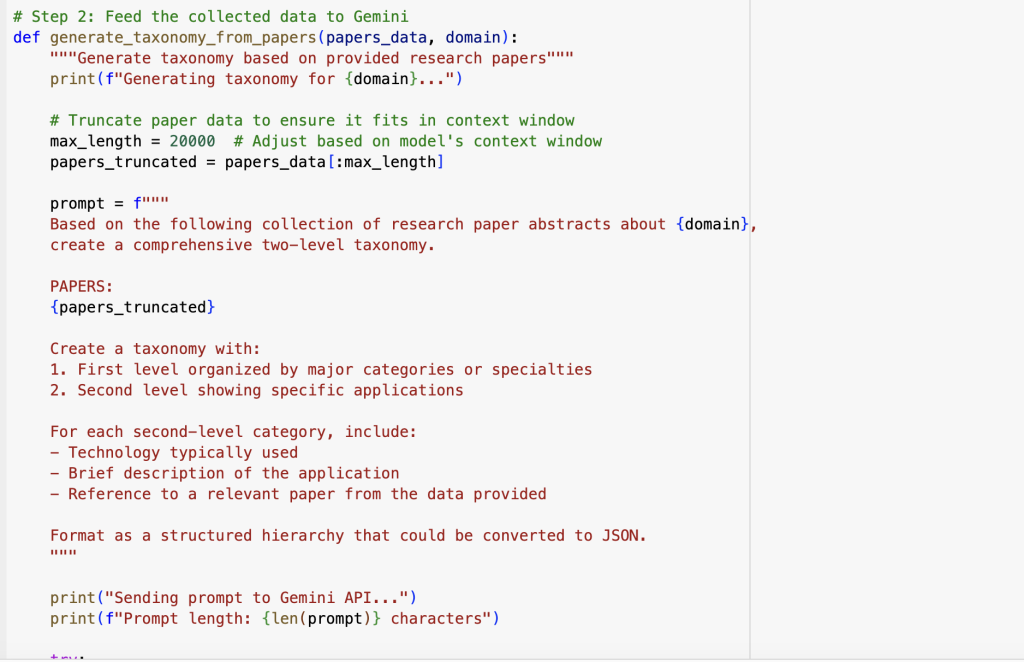
For the crucial initial data collection step, we developed a two-step process that collects real publication data and uses AI to synthesise it into a structured taxonomy:


Current Limitations and Future Enhancements
While our proof-of-concept implementation demonstrates the feasibility of automated taxonomy generation, several technical challenges remain to be addressed:
1. Deep Research API Access
The most significant limitation is access to sophisticated deep research capabilities through APIs. Current AI models like Gemini and GPT provide limited or no direct access to their web search or deep research features through their APIs. This restricts our ability to fully automate the initial data collection step.
Our current workaround—collecting papers from PubMed and then using AI to analyse them—demonstrates the concept but doesn’t fully automate the pipeline. As AI providers expand their API capabilities, we anticipate this limitation will be resolved.
2. Structured Output Consistency
Ensuring consistent, well-structured output from AI models remains challenging. Our initial implementation sometimes produces output that requires additional processing to convert to a clean JSON format.
By implementing more robust post-processing of AI outputs and exploring techniques to guide models toward more consistent, structured responses.
3. Workflow Orchestration
The full implementation would benefit from proper workflow orchestration using tools like Apache Airflow, n8n, to manage the pipeline execution.
Beyond Healthcare: Adapting the Pipeline for Different Domains
The taxonomy automation pipeline we’ve developed is inherently flexible and can be adapted to various domains beyond healthcare. The core steps—data collection, structure generation, validation, refinement, visualisation, and documentation—remain consistent, while domain-specific elements can be customised.
To adapt the pipeline to a new domain:
- Modify data sources: Replace PubMed with domain-relevant databases (e.g., arXiv for physics, IEEE for engineering)
- Adjust validation metrics: Define domain-appropriate metrics for category validation
- Customise visualisation: Adapt visual representations to domain conventions
- Refine prompts: Create domain-specific prompts that capture the unique aspects of the field.
Conclusion
Our exploration has taken us from a manually developed taxonomy of AI applications in healthcare to the vision and initial implementation of an automated taxonomy generation pipeline. This journey reflects broader trends in knowledge management—using AI tools themselves to better understand and organise information about rapidly evolving fields.
The healthcare AI taxonomy we developed provides a valuable framework for practitioners seeking to navigate the complex landscape of AI applications in medicine. By organising applications by speciality and providing information about technologies, clinical implementations, and research examples, it creates an accessible entry point for clinicians interested in understanding how AI might impact their practice.
Our current implementation represents the first iteration of this vision—a proof of concept that demonstrates the feasibility of using AI to analyse publication data and generate structured taxonomies. While still limited compared to our manually created taxonomy, it shows promising capabilities in extracting meaningful categories, identifying technologies, and referencing relevant literature.
The future development of this pipeline holds exciting possibilities. As AI capabilities continue to advance—particularly in terms of API access to deep research features and larger context windows—we anticipate that automated taxonomy generation will become increasingly sophisticated, comprehensive, and accurate.
Catch the latest version of this article over on Medium.com. Hit the button below to join our readers there.