How to use GPT API to export a research graph from PDF publications
Unveiling the Structure Within — Extracting Research Entities and Relationships
Introduction
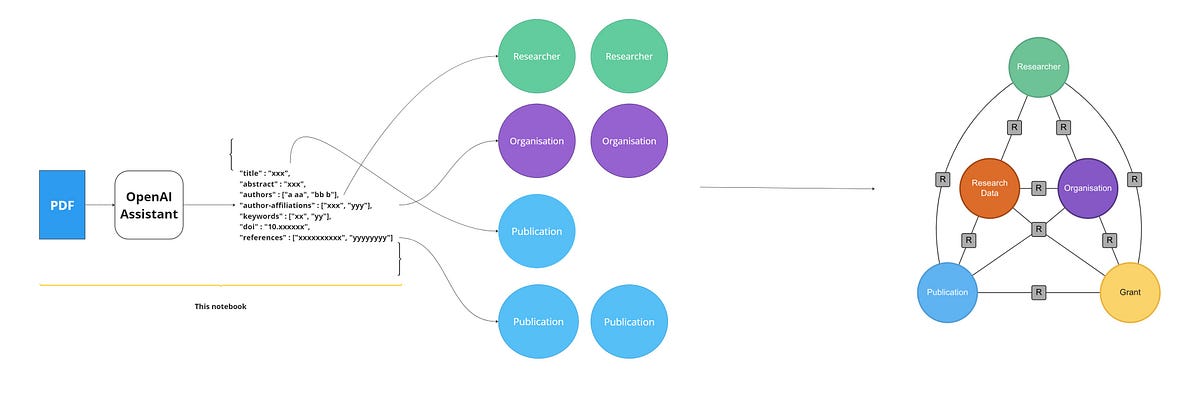
Research Graph is a structured representation of research objects that captures information about entities and the relationships between Researcher, Organisation, Publication, Grant and Research Data. Currently, the publications are available as PDF files, and due to the free-form text, it is difficult to parse a PDF file to extract structured information. In this article, we will try to create a research graph from a PDF of a publication by extracting relevant information from the text and organising it into a graph structure using OpenAI.
OpenAI
In this work, we use OpenAI API and the new Assistant functionality of GPT (Currently in Beta) to transform PDF documents to a set of structured JSON files based on Research Graph Schema.
Assistants API
The Assistants API allows you to build Artificial Intelligence (AI) assistants within your applications. An assistant can answer user questions by using models, tools, and information per predetermined guidelines. It is a beta API that is actively under development. Using the assistants API, we can use the OpenAI-hosted tools like code interpreter and knowledge retrieval. For this blog post, we will focus on knowledge retrieval.
Knowledge Retrieval
Sometimes, we need the AI model to answer queries based on unknown knowledge, like user-provided documents or sensitive information. We can augment the model with this information using the assistant APIs knowledge retrieval tool. We can upload files to the assistant, and it automatically chunks the documents and creates and stores embeddings to implement vector search on the data.
Example
For our example, we will upload PDF files of publications to the OpenAI assistant and the knowledge retrieval tool to get a JSON output of the graph schema for the given publications. The publication we used for this example can be accessed from the following link.
Step 1
Read the input path where the Publications PDFs are stored and the output path where the JSON output will be stored.
import configparser
config = configparser.ConfigParser()
config.read('{}/config.ini'.format(current_path))
input_path = config['DEFAULT']['Input-Path']
output_path = config['DEFAULT']['Output-Path']
debug = config['DEFAULT']['Debug']Step 2
Get all the PDF files from the Input path.
onlyfiles = [f for f in os.listdir(input_path) if os.path.isfile(os.path.join(input_path, f))]Step 3
Then, we need to initialise the assistant to use the Knowledge retrieval tool. To do this, we need to specify the type of tools in the API for “retrieval”. We also specify the instructions for the assistant and the OpenAI model to be used.
my_file_ids = []
if client.files.list().data==[]:
for f in onlyfiles:
file = client.files.create(
file=open(input_path + f, "rb"),
purpose='assistants'
)
my_file_ids.append(file.id)
# Add the file to the assistant
assistant = client.beta.assistants.create(
instructions = "You are a publication database support chatbot. Use pdf files uploaded to best respond to user queries in JSON.",
model = "gpt-4-1106-preview",
tools = [{"type": "retrieval"}],
# Do not attach all files to the assistant, otherwise, it will mismatch the answers even though specify file ID in query messages.
# We will attach to each message instead
)Step 4
Then, we specify the information we need to extract from the publications files and pass them as user queries to the assistant. After experimenting with the assistant instructions, we found that requesting JSON format in each user message generates the most consistent output.
user_msgs = ["Print the title of this paper in JSON",
"Print the authors of this paper in JSON",
"Print the abstract section of the paper in JSON",
"Print the keywords of the paper in JSON",
"Print the DOI number of the paper in JSON",
"Print the author affiliations of the paper in JSON",
"Print the reference section of the paper in JSON"]Step 5
The next is to pass the queries to the assistant to generate the output. We need to create an individual thread object for each user query, which contains the query as a user message. Then, we run the thread and retrieve the assistant’s answer.
all_results = []
for i in my_file_ids:
print('\n#####')
# the JSON result it can extract and parse, hopefully
file_result = {}
for q in user_msgs:
# create thread, user message and run objects for each query
thread = client.beta.threads.create()
msg = client.beta.threads.messages.create(
thread_id=thread.id,
role="user",
content=q,
file_ids=[i] # specify the file/publication we want to extract from
)
print('\n',q)
run = client.beta.threads.runs.create(
thread_id=thread.id,
assistant_id=assistant.id,
additional_instructions="If answer cannot be found, print 'False'" # not very useful at the time of this presentation
)
# checking run status by retrieving updated object each time
while run.status in ["queued",'in_progress']:
print(run.status)
time.sleep(5)
run = client.beta.threads.runs.retrieve(
thread_id=thread.id,
run_id=run.id
)
# usually a rate limit error
if run.status=='failed': logging.info("Run failed: ", run)
if run.status=='completed':
print("<Complete>")
# extract updated message object, this includes user messages
messages = client.beta.threads.messages.list(
thread_id=thread.id
)
for m in messages:
if m.role=='assistant':
value = m.content[0].text.value # get the text response
if "json" not in value:
if value=='False':logging.info("No answer found for ", str(q))
else:
logging.info("Not JSON output, maybe no answer found in the file or model is outdated: ", str(value))
else:
# clean the response and try to parse as json
value = value.split("```")[1].split('json')[-1].strip()
try:
d = json.loads(value)
file_result.update(d)
print(d)
except Exception as e:
logging.info(f"Query {q} \nFailed to parse string to JSON: ", str(e))
print(f"Query {q} \nFailed to parse string to JSON: ", str(e))
all_results.append(file_result)The JSON output generated for the above publication file is:
[{'title': 'Dodes (diagnostic nodes) for Guideline Manipulation',
'authors': [{'name': 'PM Putora',
'affiliation': 'Department of Radiation-Oncology, Kantonsspital St. Gallen, St. Gallen, Switzerland'},
{'name': 'M Blattner',
'affiliation': 'Laboratory for Web Science, Zürich, Switzerland'},
{'name': 'A Papachristofilou',
'affiliation': 'Department of Radiation Oncology, University Hospital Basel, Basel, Switzerland'},
{'name': 'F Mariotti',
'affiliation': 'Laboratory for Web Science, Zürich, Switzerland'},
{'name': 'B Paoli',
'affiliation': 'Laboratory for Web Science, Zürich, Switzerland'},
{'name': 'L Plasswilma',
'affiliation': 'Department of Radiation-Oncology, Kantonsspital St. Gallen, St. Gallen, Switzerland'}],
'Abstract': {'Background': 'Treatment recommendations (guidelines) are commonly represented in text form. Based on parameters (questions) recommendations are defined (answers).',
'Objectives': 'To improve handling, alternative forms of representation are required.',
'Methods': 'The concept of Dodes (diagnostic nodes) has been developed. Dodes contain answers and questions. Dodes are based on linked nodes and additionally contain descriptive information and recommendations. Dodes are organized hierarchically into Dode trees. Dode categories must be defined to prevent redundancy.',
'Results': 'A centralized and neutral Dode database can provide standardization which is a requirement for the comparison of recommendations. Centralized administration of Dode categories can provide information about diagnostic criteria (Dode categories) underutilized in existing recommendations (Dode trees).',
'Conclusions': 'Representing clinical recommendations in Dode trees improves their manageability handling and updateability.'},
'Keywords': ['dodes',
'ontology',
'semantic web',
'guidelines',
'recommendations',
'linked nodes'],
'DOI': '10.5166/jroi-2-1-6',
'references': [{'ref_number': '[1]',
'authors': 'Mohler J Bahnson RR Boston B et al.',
'title': 'NCCN clinical practice guidelines in oncology: prostate cancer.',
'source': 'J Natl Compr Canc Netw.',
'year': '2010 Feb',
'volume_issue_pages': '8(2):162-200'},
{'ref_number': '[2]',
'authors': 'Heidenreich A Aus G Bolla M et al.',
'title': 'EAU guidelines on prostate cancer.',
'source': 'Eur Urol.',
'year': '2008 Jan',
'volume_issue_pages': '53(1):68-80',
'notes': 'Epub 2007 Sep 19. Review.'},
{'ref_number': '[3]',
'authors': 'Fairchild A Barnes E Ghosh S et al.',
'title': 'International patterns of practice in palliative radiotherapy for painful bone metastases: evidence-based practice?',
'source': 'Int J Radiat Oncol Biol Phys.',
'year': '2009 Dec 1',
'volume_issue_pages': '75(5):1501-10',
'notes': 'Epub 2009 May 21.'},
{'ref_number': '[4]',
'authors': 'Lawrentschuk N Daljeet N Ma C et al.',
'title': "Prostate-specific antigen test result interpretation when combined with risk factors for recommendation of biopsy: a survey of urologist's practice patterns.",
'source': 'Int Urol Nephrol.',
'year': '2010 Jun 12',
'notes': 'Epub ahead of print'},
{'ref_number': '[5]',
'authors': 'Parmelli E Papini D Moja L et al.',
'title': 'Updating clinical recommendations for breast colorectal and lung cancer treatments: an opportunity to improve methodology and clinical relevance.',
'source': 'Ann Oncol.',
'year': '2010 Jul 19',
'notes': 'Epub ahead of print'},
{'ref_number': '[6]',
'authors': 'Ahn HS Lee HJ Hahn S et al.',
'title': 'Evaluation of the Seventh American Joint Committee on Cancer/International Union Against Cancer Classification of gastric adenocarcinoma in comparison with the sixth classification.',
'source': 'Cancer.',
'year': '2010 Aug 24',
'notes': 'Epub ahead of print'},
{'ref_number': '[7]',
'authors': 'Rami-Porta R Goldstraw P.',
'title': 'Strength and weakness of the new TNM classification for lung cancer.',
'source': 'Eur Respir J.',
'year': '2010 Aug',
'volume_issue_pages': '36(2):237-9'},
{'ref_number': '[8]',
'authors': 'Sinn HP Helmchen B Wittekind CH.',
'title': 'TNM classification of breast cancer: Changes and comments on the 7th edition.',
'source': 'Pathologe.',
'year': '2010 Aug 15',
'notes': 'Epub ahead of print'},
{'ref_number': '[9]',
'authors': 'Paleri V Mehanna H Wight RG.',
'title': "TNM classification of malignant tumours 7th edition: what's new for head and neck?",
'source': 'Clin Otolaryngol.',
'year': '2010 Aug',
'volume_issue_pages': '35(4):270-2'},
{'ref_number': '[10]',
'authors': 'Guarino N.',
'title': 'Formal Ontology and Information Systems',
'source': '1998 IOS Press'},
{'ref_number': '[11]',
'authors': 'Uschold M Gruniger M.',
'title': 'Ontologies: Principles Methods and Applications.',
'source': 'Knowledge Engineering Review',
'year': '1996',
'volume_issue_pages': '11(2)'},
{'ref_number': '[12]',
'authors': 'Aho A Garey M Ullman J.',
'title': 'The Transitive Reduction of a Directed Graph.',
'source': 'SIAM Journal on Computing',
'year': '1972',
'volume_issue_pages': '1(2): 131–137'},
{'ref_number': '[13]',
'authors': 'Tai K',
'title': 'The tree-to-tree correction problem.',
'source': 'Journal of the Association for Computing Machinery (JACM)',
'year': '1979',
'volume_issue_pages': '26(3):422-433'}]}]Step 6
File objects and assistant object needs to be cleaned up because they cost money in ‘retrieval’ mode. Also, it is a good coding practice.
for f in client.files.list().data:
client.files.delete(f.id)# Retrieve and delete running assistants
my_assistants = client.beta.assistants.list(
order="desc",
)
for a in my_assistants.data:
response = client.beta.assistants.delete(a.id)
print(response)Step 7
The next step is to generate a graph visualisation using the Python Networkx package.
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph()
node_colors = []
key = "jroi/" + all_results[0]['title']
G.add_nodes_from([(all_results[0]['title'], {'doi': all_results[0]['DOI'], 'title': all_results[0]['title'], 'source': 'jroi', 'key': key})])
node_colors.append('#4ba9dc')
for author in all_results[0]['authors']:
key = "jroi/" + author['name']
G.add_nodes_from([(author['name'], {'key': key, 'local_id': author['name'], 'full_name': author['name'], 'source': 'jroi'})])
G.add_edge(all_results[0]['title'], author['name'])
node_colors.append('#63cc9e')
for reference in all_results[0]['references']:
key = "jroi/" + reference['title']
G.add_nodes_from([(reference['title'].split('.')[0][:25] + '...', {'title': reference['title'], 'source': 'jroi', 'key': key})])
G.add_edge(all_results[0]['title'], reference['title'].split('.')[0][:25] + '...')
node_colors.append('#4ba9dc')
pos = nx.spring_layout(G)
labels = nx.get_edge_attributes(G, 'label')
nx.draw(G, pos, with_labels=True, node_size=1000, node_color=node_colors, font_size=7, font_color='black')
nx.draw_networkx_edge_labels(G, pos, edge_labels=labels)
plt.savefig("graph_image.png")
plt.show()The graph visualisation is as below:
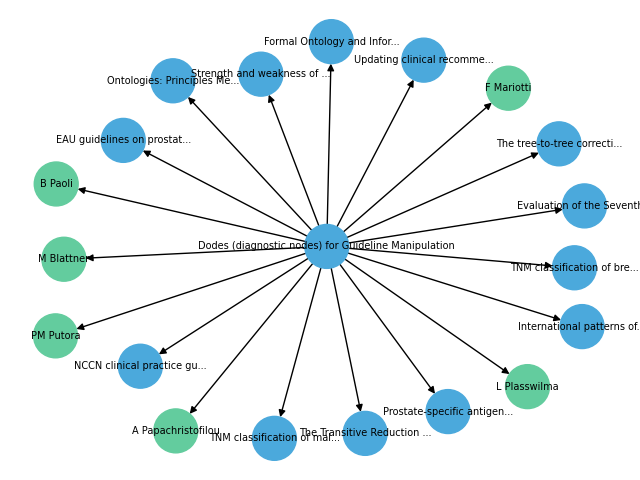
Note: Please note that the output structure generated by OpenAI may be different for different executions. So you may need to update the above code according to that structure.
Conclusion
In conclusion, leveraging the GPT API to extract a Research Graph from PDF publications offers a powerful and efficient solution for researchers and data analysts. This workflow streamlines the process of converting PDF publications into structured and accessible Research Graphs. But we must also pay careful attention to the inconsistencies of the responses generated by Large Language Models (LLMs). Over time, accuracy and relevance may be further enhanced by routinely updating and improving the extraction model.