How to use new Research Graph API? Finding Researchers and Organisations
Introduction
In today's data-driven research landscape, the ability to connect, query, and extract insights from interconnected research data is more valuable than ever.
In this blog post, we'll explore how to build REST APIs using Flask and Neo4j to expose research network data in an accessible way. We'll dive deep into two complementary APIs: one for searching researchers and another for exploring organisations, both implemented with Python, Flask, and Neo4j's Cypher query language.
The Research Knowledge Graph
Before diving into the implementation details, it's important to understand the data model we're working with. The Research Graph meta model (available at https://researchgraph.org/schema/) provides a comprehensive structure for representing research entities and their relationships.
In our knowledge graph implementation, we focus on four primary node types:
- Researchers: Academic professionals with attributes like names, identifiers (ORCID), and affiliations
- Publications: Research outputs like papers, articles, and books
- Organisations: Research institutions, universities, and other entities
- Grants: Research funding awarded to researchers and organisations
These nodes are connected through various relationships that represent real-world connections, such as:
- Researchers PUBLISH publications
- Researchers are AFFILIATED WITH organisations
- Researchers RECEIVE grants
- Organisations ADMINISTER grants
Let's examine how these relationships are represented in our Cypher queries:
// Researcher-Publication relationships
(r:researcher)-[pub_rel:orcid|crossref|pubmed]->(p:publication)
// Researcher-Organization relationships
(r:researcher)-[:orcid]->(o:organisation)
// Researcher-Grant relationships
(r)-[grant_rel:researchgraph|arc|orcid]->(g:grant)
// Organization-Grant relationships
(o)-[:wikidata|arc]->(g1:grant)
The power of representing research data as a graph lies in the ability to traverse these connections efficiently and uncover insights that would be difficult to extract from traditional data models. Our APIs leverage this interconnectedness to provide rich context around each entity.
Setting Up the Foundation
Our API implementation begins with setting up a solid foundation for logging, configuration, and database connectivity. Let's examine how we handle logging configuration:
def setup_logging():
# Use current working directory as base
log_dir = os.path.join(os.getcwd(), 'logs')
os.makedirs(log_dir, exist_ok=True)
log_file = os.path.join(log_dir, 'application.log')
logging.basicConfig(
level=logging.INFO,
format='%(asctime)s - %(name)s - %(levelname)s - %(message)s',
handlers=[
logging.FileHandler(log_file),
logging.StreamHandler() # Also log to console
]
)
return logging.getLogger(__name__)
This setup ensures that all application events are captured both in a log file and in the console, which is essential for debugging and monitoring the application in production.
For configuration management, we use Python's built-in ConfigParser module to read settings from a config.ini file:
def read_config():
config = configparser.ConfigParser()
config_path = os.path.join(os.getcwd(), 'config.ini')
config.read(config_path)
return config
This approach makes our application configurable without requiring code changes, which is particularly valuable when deploying in different environments.
Neo4j Connection Management
The core of our APIs is the Neo4jConnection class, which encapsulates all interactions with the Neo4j database:
class Neo4jConnection:
def __init__(self, uri, user, password, result_limit=5, max_distance=3):
try:
self._driver = GraphDatabase.driver(uri, auth=(user, password))
self.result_limit = result_limit
self.max_distance = max_distance
except Exception as e:
logger.error(f"Failed to connect to Neo4j: {str(e)}")
raise
This class handles connection establishment, query execution, and result processing. By encapsulating these functions, we maintain a clean separation of concerns and make our Flask routes more focused on HTTP-specific logic.
The connection is initialised using configuration parameters, which we get from config.ini file:
def init_neo4j_connection():
try:
config = read_config()
neo4j_uri = config.get('Neo4j', 'uri', fallback='bolt://localhost:7687')
neo4j_user = config.get('Neo4j', 'user', fallback='neo4j')
neo4j_password = config.get('Neo4j', 'password')
result_limit = config.getint('Neo4j', 'result_limit', fallback=5)
max_distance = config.getint('Neo4j', 'max_distance', fallback=3)
return Neo4jConnection(neo4j_uri, neo4j_user, neo4j_password, result_limit, max_distance)
except Exception as e:
logger.critical(f"Failed to initialize Neo4j connection: {str(e)}")
raiseThe Researcher API
Our researcher API provides a way to search for researchers by name and retrieve detailed information about them, including their publications, grants, and organisational affiliations.
Here's the Flask route that exposes this functionality:
@app.route('/api/researchers', methods=['GET'])
def search_researchers_api():
name = request.args.get('name', '')
# Validate input
if not name:
logger.warning("Search attempted without name parameter")
return jsonify({
"error": "Name parameter is required",
"status": 400
}), 400
try:
# Search for researchers
researchers = neo4j_conn.search_researchers(name)
return jsonify({
"status": "success",
"count": len(researchers),
"researchers": researchers
})
except Exception as e:
logger.error(f"Error processing researcher search: {str(e)}")
return jsonify({
"error": "An error occurred while processing the request",
"status": 500
}), 500
The core of the researcher search functionality is the Cypher query in the search_researchers method:
def search_researchers(self, name):
query = """
MATCH (r:researcher)
WHERE r.orcid IS NOT NULL
WITH r, apoc.text.distance(
toLower(r.full_name),
toLower($name)
) AS edit_distance
WHERE edit_distance <= 3
OPTIONAL MATCH (r)-[pub_rel:orcid|crossref|pubmed]->(p:publication)
OPTIONAL MATCH (r)-[:orcid]->(o:organisation)
WITH
r,
edit_distance,
COUNT(DISTINCT p) AS publication_count,
COLLECT(DISTINCT o.name) AS affiliations
OPTIONAL MATCH (r)-[grant_rel:researchgraph|arc|orcid]->(g:grant)
RETURN
r.orcid AS orcid,
r.full_name AS researcher_name,
r.key AS key,
r.source AS source,
r.local_id AS local_id,
r.last_updated AS last_updated,
r.full_name AS full_name,
r.first_name AS first_name,
r.last_name AS last_name,
r.url AS url,
r.scopus_author_id AS scopus_author_id,
publication_count,
COUNT(DISTINCT g) AS funding_count,
affiliations,
edit_distance AS distance
ORDER BY edit_distance ASC, publication_count + COUNT(DISTINCT g) DESC
LIMIT $limit
"""This query uses advanced fuzzy matching techniques:
- It finds researchers who have an ORCID identifier (ensuring reliable identification)
- It uses the Levenshtein distance algorithm (via apoc.text.distance) to match names with some tolerance for spelling variations
- It restricts results to names within an edit distance of 3 (allowing for minor typos and variations) and handles accents in names.
- It traverses relationships to count publications and grants
- It collects all organisational affiliations
- It orders results first by edit distance (closer matches first) and then by the combined publication and grant counts. This sorting ensures that users see the most relevant results first.
- It returns the edit distance in the results, allowing clients to gauge match quality
- It limits results based on the configured limit
The Organisation API
The organisation API complements the researcher API by providing a way to search for research organisations and retrieve information about them, including affiliated researchers, publications, and grants.
def search_organizations(self, name):
query = """
MATCH (o:organisation)
WHERE o.name IS NOT NULL
WITH o, apoc.text.distance(
toLower(o.name),
toLower($name)
) AS edit_distance
WHERE edit_distance <= $max_distance
// Find affiliated researchers
OPTIONAL MATCH (o)<-[:orcid]-(r:researcher)
// Find publications through researchers
OPTIONAL MATCH (r)-[:orcid|crossref|pubmed]->(p:publication)
// Find grants directly connected to organization
OPTIONAL MATCH (o)-[:wikidata|arc]->(g1:grant)
// Find grants through researchers
OPTIONAL MATCH (r)-[:orcid]->(g2:grant)
WITH
o,
edit_distance,
COUNT(DISTINCT r) AS researcher_count,
COUNT(DISTINCT p) AS publication_count,
COUNT(DISTINCT g1) + COUNT(DISTINCT g2) AS funding_count
RETURN
o.name AS organisation_name,
o.key AS key,
o.source AS source,
o.local_id AS local_id,
o.last_updated AS last_updated,
o.url AS url,
o.grid AS grid,
o.doi AS doi,
o.ror AS ror,
o.isni AS isni,
o.wikidata AS wikidata,
o.country AS country,
o.city AS city,
o.latitude AS latitude,
o.longitude AS longitude,
researcher_count,
publication_count,
funding_count,
edit_distance AS distance
ORDER BY edit_distance ASC, researcher_count DESC
LIMIT $limit
"""
This query:
- Uses the APOC library's text distance function to find organisations with names similar to the search term
- Applies a maximum edit distance filter to limit results to reasonable matches
- Traverses multiple relationship paths to count affiliated researchers, publications, and grants
- Orders results first by edit distance (closer matches first) and then by researcher count
- Returns comprehensive organisation metadata, including identifiers and location information
Error Handling and Validation
Both APIs implement robust error handling and input validation:
# Validate input
if not name:
logger.warning("Search attempted without name parameter")
return jsonify({
"error": "Name parameter is required",
"status": 400
}), 400
For exception handling:
try:
# Search for organizations
organizations = neo4j_conn.search_organizations(name)
return jsonify({
"status": "success",
"count": len(organizations),
"organizations": organizations
})
except Exception as e:
logger.error(f"Error processing organization search: {str(e)}")
return jsonify({
"error": "An error occurred while processing the request",
"status": 500
}), 500
This approach ensures that:
- Invalid requests are rejected with clear error messages
- Unexpected errors are caught and logged for debugging
- The API maintains a consistent response format regardless of success or failure
Accessing the APIs
Using Postman
Postman provides an excellent way to explore and test our APIs. Here's how to set up a request to search for researchers:
- Create a new GET request in Postman
- Set the URL to https://researchgraph.cloud/api/national-graph/researchers
- Add a parameter called "name" and add a name value
- Go to Authorization tab and set Auth type as Basic Auth.
- Enter your username and password
- Send the request and examine the JSON response
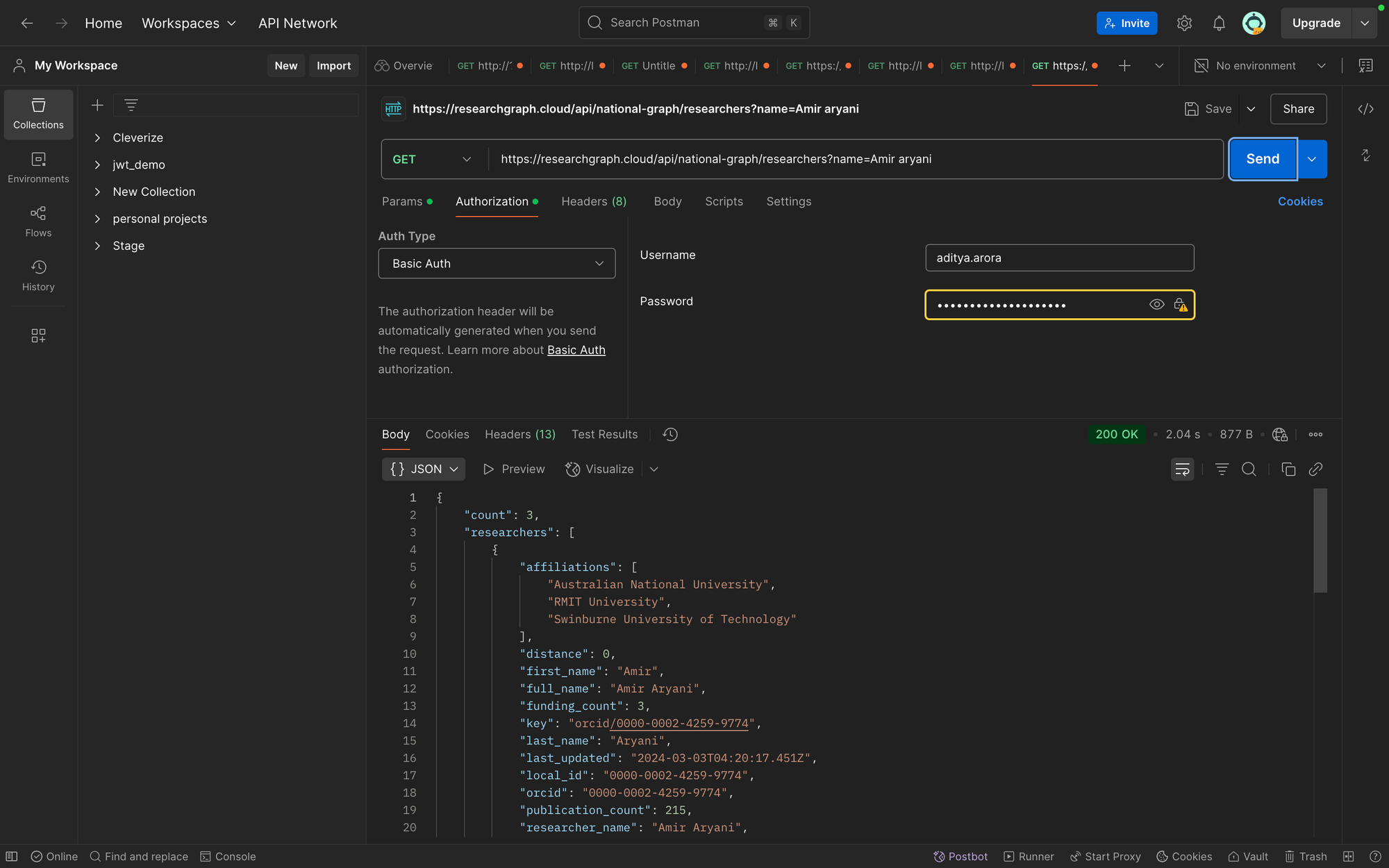
For organisations, use:
Postman allows you to save these requests, create collections, and even generate code snippets for various programming languages.

Using Python
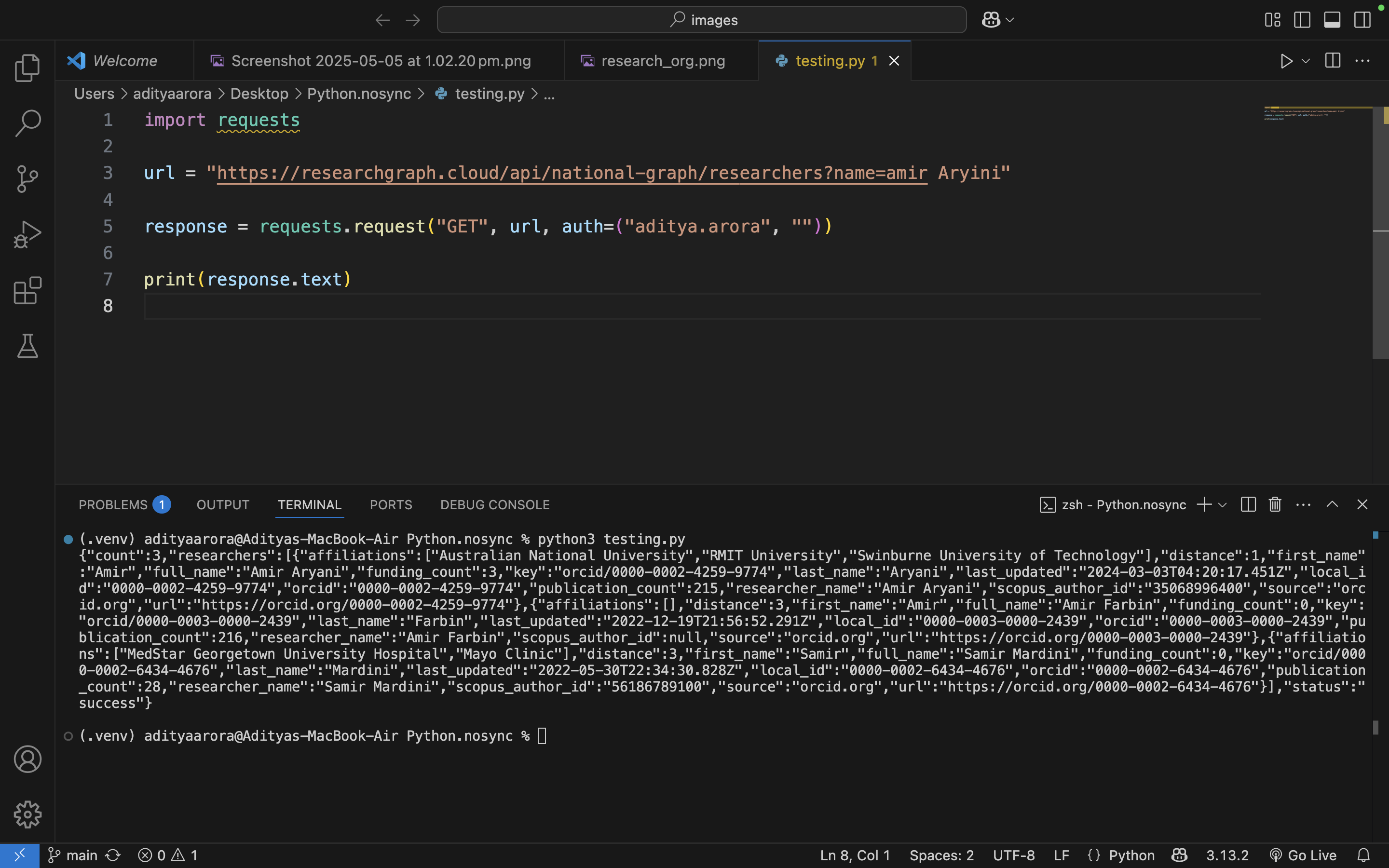
Here's a simple Python client that can query researcher API:
import requests
url = "https://researchgraph.cloud/api/rgf/researchers?name=Amir aryani"
response = requests.request("GET", url, auth=("aditya.arora", "password"))
print(response.text)
Conclusion
In this blog post, we've explored how to build powerful research knowledge graph APIs using Flask and Neo4j. By leveraging the natural graph structure of research data, we can provide rich, contextual information about researchers and organisations that would be difficult to extract from traditional databases.
The combination of Flask's simplicity, Neo4j's graph query capabilities, and Python's ecosystem makes for a powerful toolkit for building research information systems. Whether you're building applications for academic institutions, funding agencies, or research collaboration platforms, these APIs provide a solid foundation for accessing and analysing research network data.
By exposing this data through well-designed REST APIs, we make it accessible to a wide range of client applications, from web interfaces to data analysis scripts, enabling new insights and applications in the research information space.